GROE Camelina: Genomic Research for Oil Enhancement in Camelina
2017 Awardee from The Biological and Environmental Research Office of the Department of Energy
Project Website: https://camregbase.org/groe
Principal Institution: Michigan State University
Partner Institutions: North Carolina State University, Yield10 Bioscience
Our research plan aims to establish the non-food oilseed crop plant, Camelina sativa, as a commercially viable, dedicated biofuels and bioproducts feedstock. We will focus on improving seed and oil yields by employing an integrated genetic and metabolic systems approach to increase the rates of photosynthetic CO2 capture and conversion to triacylglycerols (TAGs). Camelina has the advantages of low agronomic inputs and natural resistance to biotic and abiotic stresses relative to other oilseed crops, and Camelina oil-based blends have been tested and approved as liquid transportation fuels. Camelina also benefits as a synthetic biology platform from a fully sequenced genome, well-established molecular genetic tools, and numerous resources available from its close relative, Arabidopsis thaliana. Despite these advantages, the major limitation in widespread adoption of Camelina as an industrial oilseed crop is its modest oil yield. Our proposal will address yield directly by employing a tissue-specific and whole-plant systems approach to identify the major regulatory mechanisms that limit:
1) Carbon fixation in photosynthetically active source tissues (leaves)
2) The transport of fixed carbon from source to sink tissues (seeds)
3) The allocation of fixed carbon to TAG production.
The limiting factors identified in our analyses will be individually validated and combined using multi-gene stacking and genome editing technologies to engineer Camelina. Our overall objective is to achieve up to a 300% increase per hectare in oil production, thereby meeting the yield and cost targets of a competitive biofuels and bioproducts crop while retaining its advantages for growth in marginal environments.
Our research program builds on the established expertise and collaborative record of the co-PIs to develop an integrated systems-level strategy to accelerate the development of a pipeline of potential yield traits. To this end, we will combine predictive metabolic flux modeling, quantitative proteomics, and the identification of transcriptional regulatory networks with the optimization of multi-gene and genome editing synthetic biology tools to enable the efficient analysis of stacked-traits for improved seed yield and TAG production in Camelina. Newly discovered traits from the co-PI laboratories that positively impact CO2 capture, transport and allocation will serve as the initial foundation of our strategy by providing immediate candidates for multi-gene transgenic and genome editing approaches to test for additive or synergistic effects on oil yields. These traits will also serve as the platforms for performing tissue-specific and whole-plant metabolic flux and transcriptional regulatory network analyses for the development of dynamic metabolic and gene regulatory models. These predictive models will accelerate the identification of additional metabolic bottlenecks, transcriptional regulators and/or post-translational mechanisms that control seed yields. The control points identified using our systems-level models provide novel targets for engineering and yield analysis individually and in combination with our proven yield traits. Our studies will be guided by an integrated workflow plan for optimized discovery in which output from our predictive, systems-level models inform the selection and analysis of novel trait combinations. Likewise, information from our trait analysis will provide input for the further refinement of our predictive models. This results in a highly integrated and robust research plan that maximizes the potential for achieving our seed and oil yield goals.
Development of Tissue-Specific and Whole-Plant Metabolic Network Models to Identify
Additional Control Points that Limit Seed Oil Production
(Lead PI: Dr. Yair Shachar-Hill)
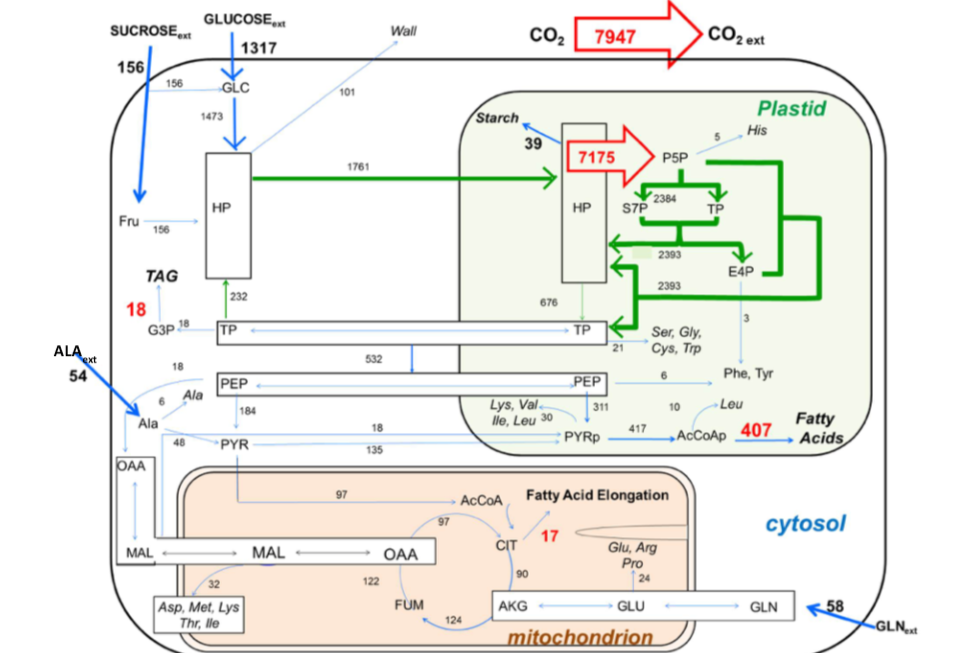 Caption (Figure left): Metabolic Flux Map showing the rates of flow (nanomoles per embryo per day) through
central metabolism. Embryos take up glucose, sucrose alanine and glutamine (bold blue
arrows) and convert them to TAG, Protein, and Starch. The total production of CO2
and flux into the OPPP are shown as red arrows. Names of metabolites abbreviated according
to standard usage, except: HP, hexose phosphates, TP, triose phosphates.
Caption (Figure left): Metabolic Flux Map showing the rates of flow (nanomoles per embryo per day) through
central metabolism. Embryos take up glucose, sucrose alanine and glutamine (bold blue
arrows) and convert them to TAG, Protein, and Starch. The total production of CO2
and flux into the OPPP are shown as red arrows. Names of metabolites abbreviated according
to standard usage, except: HP, hexose phosphates, TP, triose phosphates.
Workflow plan:
To identify metabolic and/or transport processes that restrict seed yields, we will analyze the flow of carbon from CO2 fixation to the synthesis of seed storage compounds; this information will inform transcriptomic and proteomic analyses to pinpoint molecular targets for genetic engineering. Metabolic flux analysis tools will be applied at three levels: carbon assimilation and related central carbon metabolism in source leaves; the transport and allocation of photoassimilate to sink tissues; and the use of carbon for TAG synthesis in seeds. The MFA strategy will use a time course of whole plant labeling with 13/14CO2 to trace the transport and allocation of photoassimilate from leaves through the vascular tissue to sink tissues (roots, seeds, pod tissue). These data will be combined with MFA data from source and sink tissues to identify the steps and tissues (assimilation, transport, allocation) that limit the conversion of fixed carbon to TAGs. To accomplish this, we will combine labeling time course experiments in whole plants with Metabolic Control Analysis (MCA). The Shachar-Hill group has implemented dynamic labeling flux analysis methods, which enabled us to apply MCA methods to identify targets for carbon allocation control in cell wall production.
Lipid analysis of Camelina sativa seeds and cultured embryos
(Lead PI: Dr. Mike Pollard)
Reference: Pollard M, Delamarter D, Martin TM, Shachar-Hill Y. Lipid labeling from acetate or glycerol in cultured embryos of Camelina sativa seeds: A tale of two substrates. Phytochemistry. 2015 Oct;118:192-203. doi: 10.1016/j.phytochem.2015.07.021. Epub 2015 Aug 8.